Systematic uncertainties
[1]:
import pandas as pd
import seaborn as sns
from freeforestml import Variable, Process, Cut, hist, SystStack, DataStack, \
McStack, TruthStack
from freeforestml import toydata, example_style
example_style()
2023-08-02 16:40:04.927633: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2023-08-02 16:40:05.065114: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcudart.so.11.0'; dlerror: libcudart.so.11.0: cannot open shared object file: No such file or directory
2023-08-02 16:40:05.065144: I tensorflow/compiler/xla/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
2023-08-02 16:40:05.906728: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer.so.7'; dlerror: libnvinfer.so.7: cannot open shared object file: No such file or directory
2023-08-02 16:40:05.906837: W tensorflow/compiler/xla/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer_plugin.so.7'; dlerror: libnvinfer_plugin.so.7: cannot open shared object file: No such file or directory
2023-08-02 16:40:05.906849: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Cannot dlopen some TensorRT libraries. If you would like to use Nvidia GPU with TensorRT, please make sure the missing libraries mentioned above are installed properly.
[2]:
df = toydata.get()
df = df.compute()
[3]:
df = df.assign(variation="NOMINAL")
df_up = df.assign(higgs_m=df.higgs_m + 1 * df.jet_1_eta.abs(),
variation="1up")
df_down = df.assign(higgs_m=df.higgs_m - 1 * df.jet_2_eta.abs(),
variation="1down")
df = pd.concat([df, df_up, df_down], sort=False)
[4]:
p_ztt = Process(r"$Z\rightarrow\tau\tau$", range=(0, 0))
p_sig = Process(r"Signal", range=(1, 1))
p_bkg_up = Process(r"Up", lambda d: d.variation == "1up")
p_bkg_down = Process(r"Down", lambda d: d.variation == "1down")
p_ztt_nom = Process(r"$Z\rightarrow\tau\tau$", lambda d: (d.variation == "NOMINAL") & (d.fpid == 0))
p_sig_nom = Process(r"Signal", lambda d: (d.variation == "NOMINAL") & (d.fpid == 1))
p_asimov = Process(r"Asimov", selection=lambda d: (d.fpid >= 0) & (d.variation == "NOMINAL"))
[5]:
c_nominal = Cut(lambda d: d.variation == "NOMINAL")
[6]:
colors = ["windows blue", "amber", "greyish", "faded green", "dusty purple"]
s_bkg = McStack(p_ztt, p_sig, palette=sns.xkcd_palette(colors))
s_bkg_up = TruthStack(p_bkg_up,
histtype="step",
color='k',
palette=sns.xkcd_palette(colors))
s_bkg_down = TruthStack(p_bkg_down,
histtype="step",
color='k',
linestyle='--',
palette=sns.xkcd_palette(colors))
s_bkg_nom = McStack(p_ztt_nom, p_sig_nom, palette=sns.xkcd_palette(colors))
s_bkg_syst = SystStack(p_ztt, p_sig, palette=sns.xkcd_palette(colors))
s_data = DataStack(p_asimov)
[7]:
v_higgs_m = Variable(r"$m^H$", "higgs_m", "GeV")
Plot nominal
[8]:
hist(c_nominal(df), v_higgs_m, 20, [s_bkg, s_data], range=(0, 200), selection=None,
weight="weight", ratio_label="Data / SM", ratio_range=(0.1, 1.9))
None
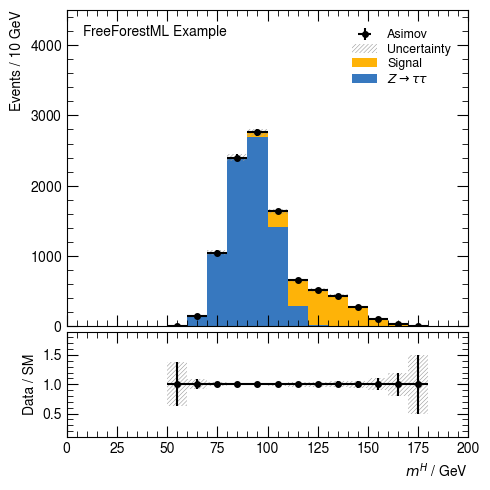
Envelop
[9]:
hist(df, v_higgs_m, 20, [s_bkg_nom, s_bkg_up, s_bkg_down], range=(0, 200), selection=None,
weight="weight", ratio_label="Up / Down", ratio_range=(0.1, 1.9))
None
/home/docs/checkouts/readthedocs.org/user_builds/freeforestml/envs/latest/lib/python3.7/site-packages/freeforestml/stack.py:246: RuntimeWarning: invalid value encountered in multiply
return bins[:-1] * 0
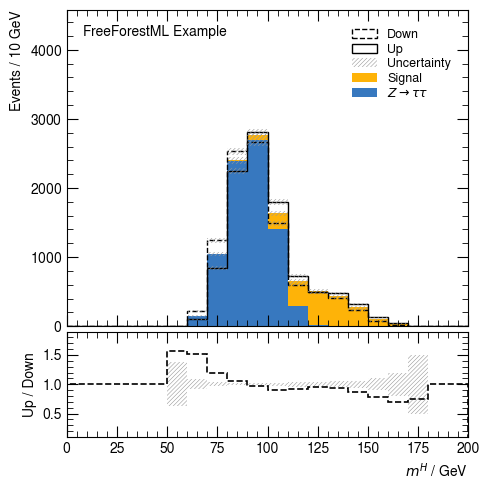
Full band
[10]:
hist(df, v_higgs_m, 20, [s_bkg_syst, s_data], range=(0, 200), selection=None,
weight="weight", ratio_label="Data / SM", ratio_range=(0.1, 1.9))
None
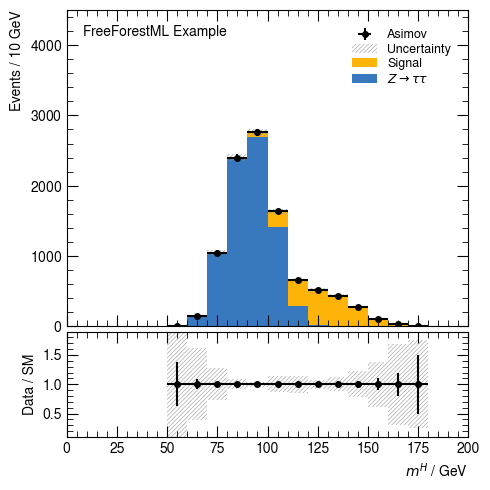
[ ]: